The way how whole blood and plasma interpolation is performed in PKIN is governed by the configuration on the Blood tab. There are two selection items:
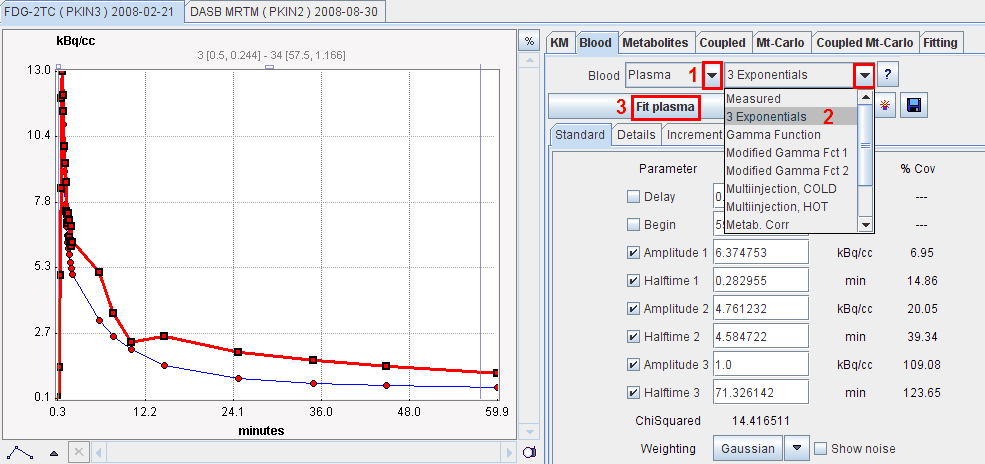
The parameters of the blood model appear in the Standard pane and can be configured for fitting purposes. Activating the Fit plasma or Fit Whole Blood button starts a fitting process which adjusts the model parameters such that the interpolation curve comes into optimal agreement with the measurements. In the example below the model has not yet been fitted so that the distinction between the measurement and the model (sum of exponentials) is clearly visible. For more information about the fitting options please refer to the fitting of kinetic models.
Blood Models
In PKIN there are different choices of such blood models.
Measured:
This is the default model, whereby the input curve is linearly interpolated between measured values. Outside the measured values the interpolation rules are as follows: The input curve is
3 Exponentials:
In this model the tail of the input curve from a starting point to be specified is replaced by a sum of up to three exponentials. The input curve before the start time is linearly interpolated as for the measured model.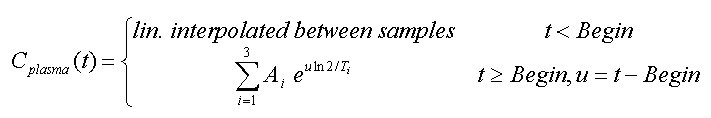
Compartment Model, 3 Eigenvalues:
This model has been developed for the FDG tracer [14]. It is the result from modeling the distribution and delivery of FDG in the circulatory system by a compartment model, and is given by 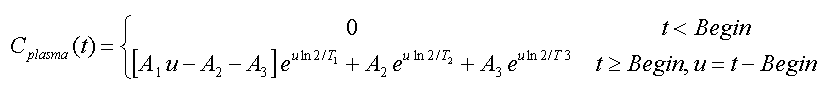
Compartment Model, 2 Eigenvalues:
This is a reduced version of the above model given by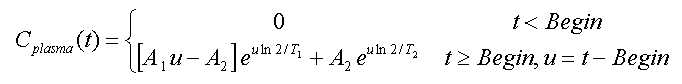
Metabolite correction (deprecated):
This model has been developed for correcting the buildup of the CO2 metabolite in 11C acetate studies [22]. The model assumes that the concentration of authentic ligand in plasma can be calculated from total blood activity by the following simple equation

where met denotes the metabolite level which is finally reached, and T represents the half-time of the exponential metabolite build-up. This model can be applied for other tracers with a similar behavior of metabolite buildup.
Note: the Metabolite correction model can not be fitted against the blood data. One way to use this metabolite correction is to determine the two parameters externally and enter them as constants in the model. An alternative is to estimate the metabolite parameters together with the kinetic parameters during a fit of the kinetic model (with the Fit blood parameters check enabled).
Bolus/Infusion optimization:
This model has been developed for the optimization of the activity ratio between an initial bolus and a subsequent infusion [23]. The aim is, that the activity level in plasma and in the tissue gets constant as soon as possible.
Multiinjection, HOT or COLD:
These models are used in combination with the model for multi-injection studies with 11C-Flumazenil as described by Delforge et al. [26]. With the HOT model, there is an analytical correction for the buildup of metabolites included. The COLD model is entirely derived from the hot input curve based on the relative doses and includes similar metabolite correction.
The Blood Delay Parameter
All standard blood models have a Delay parameter to correct for a timing error between tissue and blood data. Positive delays represent delayed blood information and hence shift the blood curves to earlier times (to the left). This parameter is only relevant for fitting of the kinetic model. Therefore, when fitting the shape of the blood curve with Fit plasma or Fit whole blood, it is automatically disabled.
However, when fitting kinetic compartment models, it is possible to fit the blood delay as an additional parameter to the parameters of the kinetic model.
Blood delay fitting
To find out the fitted blood delay, select the Blood tab after fitting. The fit value is shown, and the input curve model appears shifted accordingly.
CAUTION: When Fit blood param is enabled, all checked parameters of the input curve model are fitted. So if other parameters than Delay are also enabled, the shape of the input curve will also change!
Default Values of Model Parameters
Each model has default values. Initially, they are factory settings, but they can be re-defined by the user if he wants to establish a default configurations which is more adequate from him. Note that the configurations are valid per PMOD login, so different logins could be prepared for processing types of data requiring different initial parameter values.
|
Saves the current configuration of the model as the new default. Included are the values, the fit flags, and the restrictions of all parameters. |
|
Retrieves the default configuration of the model. |