Wu and Carson [32] aimed at making the Simplified Reference Tissue Model (SRTM) more robust for pixel-wise applications and called it Simplified Reference Tissue Model 2 (SRTM2). They noted that with SRTM k2' is calculated with each pixel TAC, although the same reference TAC is used for all pixels. Therefore they implemented a two-step approach:
The operational equation of the SRTM was re-written to allow for fixing of k2'
![]()
The three parameters R1, k2a and k'2 are estimated in step 1. In step 2 k2' is fixed, and only R1 and k2a are estimated. k2a is the apparent k2 (k2a = k2 /(1+BP)).
The binding potential can then be calculated as
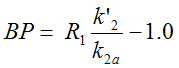
SRTM2 is based on the same assumptions as SRTM:
Note: As with the SRTM method, BP estimates from SRTM2 tend to be biased if the 1-tissue compartment model assumption does not apply. The magnitude of the bias is even larger for the SRTM2 estimates, most likely because the fixed k2' can not compensate any more a part of the model inadequacy [32].
Implementation Notes
After switching to the Simplified Ref Tissue Model 2 in PKIN a suitable reference region must be selected. For convolution with the exponentials, the reference tissue TAC is resampled on a regular grid, which can be specified by the resampling interval parameter in PKIN.
Note that k2', the transfer of tracer from the reference tissue back to the plasma, can also be fitted. After removing the fit check of k2', it remains fix at the entered value. This allows to study the bias when fixing k2'.
Abstract [32]
"The Simplified Reference Tissue Model (SRTM) produces functional images of receptor binding parameters using an input function derived from a reference region and assuming a model with one tissue compartment. Three parameters are estimated: binding potential (BP), relative delivery (R1), and the reference region clearance constant k2'. Since k'2 should not vary across brain pixels, the authors developed a two-step method (SRTM2) using a global value of k2'. Whole-brain simulations were performed using human input functions and rate constants for [18F]FCWAY, [11C]flumazenil, and [11C]raclopride, and parameter SD and bias were determined for SRTM and SRTM2. The global mean of k2' was slightly biased (2% to 6%), but the median was unbiased (<1%) and was used as the global value. Binding potential noise reductions with SRTM2 were 4% to 14%, 20% to 53%, and 10% to 30% for [18F]FCWAY, [11C]flumazenil, and [11C]raclopride, respectively, with larger reductions for shorter scans. R1 noise reduction was larger than that of BP. Simulations were also performed to assess bias when the reference and/or tissue regions followed a two-tissue compartment model. Owing to the constrained k2', SRTM2 showed somewhat larger biases due to violations of the one- compartment model assumption. These studies demonstrate that SRTM2 should be a useful method to improve the quality of neuroreceptor functional images."