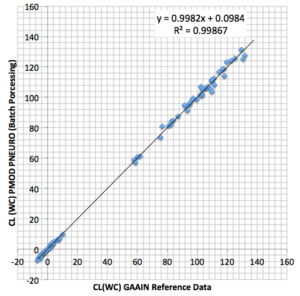
In their 2014 paper, Klunk et al. [1] highlight the need for standardization in reporting the results of amyloid PET. Their “Centiloid” methodology allows calculating a calibrated measure of the amyloid load for different tracers if data are acquired with a consistent acquisition/reconstruction methodology.
The PNEURO tool is ideally suited for this type of image processing. It includes the required brain region definitions and supports all processing steps for analysing amyloid data according to the “Centiloid” procedure.
The Application Note accessible below reviews the “Centiloid” principles and describes how a PET center can establish the “Centiloid” methodology for data of a particular amyloid tracer acquired and reconstructed with their in-house protocols.
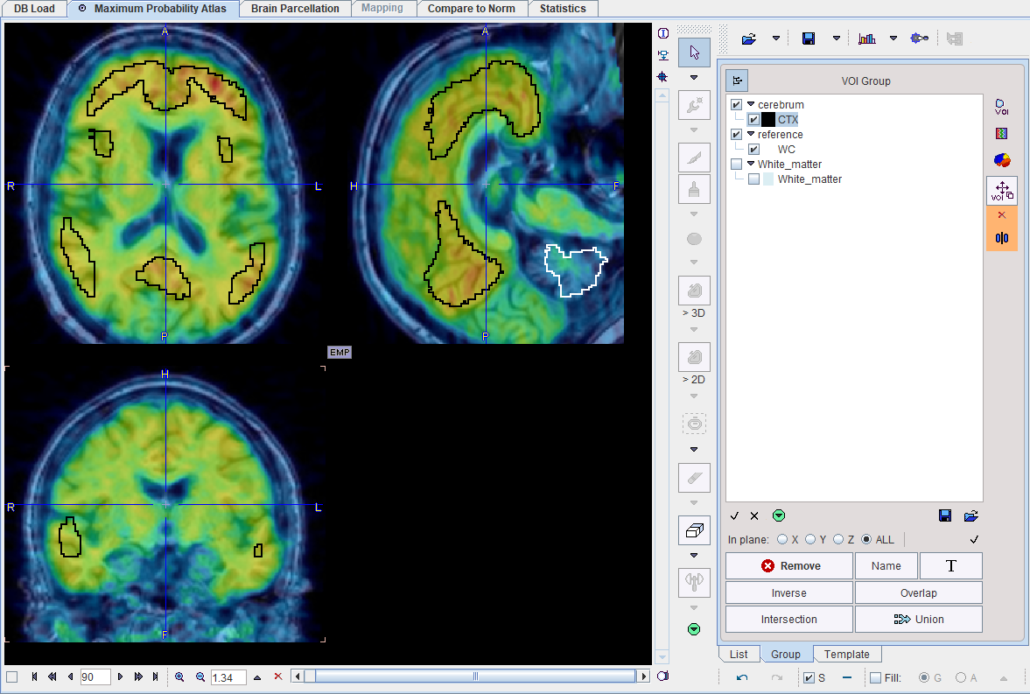
Centiloid regions on top of a PiB image acquired 50-70 min p.i. Note the slightly unexpected shape and location of the cortex region, which was empirically derived by Klunk et al. [1].
- Klunk WE, Koeppe RA, Price JC, Benzinger TL, Devous MD Sr, Jagust WJ, Johnson KA, Mathis CA, Minhas D, Pontecorvo MJ, Rowe CC, Skovronsky DM, Mintun MA. The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015; 11(1):1-15.e1-4. DOI
PMOD is a software FOR RESEARCH USE ONLY (RUO) and must not be used for diagnosis or treatment of patients.